Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes
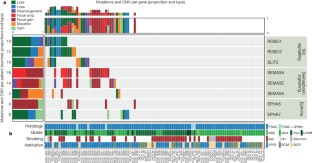
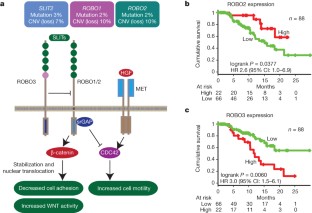
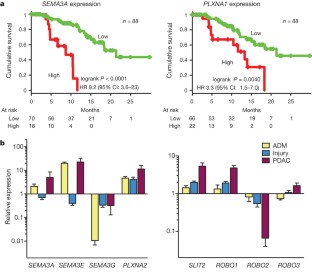
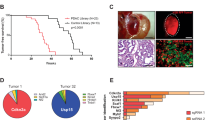
Pancreatic cancer is a highly lethal malignancy with few effective therapies. We performed exome sequencing and copy number analysis to define genomic aberrations in a prospectively accrued clinical cohort (n = 142) of early (stage I and II) sporadic pancreatic ductal adenocarcinoma. Detailed analysis of 99 informative tumours identified substantial heterogeneity with 2,016 non-silent mutations and 1,628 copy-number variations. We define 16 significantly mutated genes, reaffirming known mutations (KRAS, TP53, CDKN2A, SMAD4, MLL3, TGFBR2, ARID1A and SF3B1), and uncover novel mutated genes including additional genes involved in chromatin modification (EPC1 and ARID2), DNA damage repair (ATM) and other mechanisms (ZIM2, MAP2K4, NALCN, SLC16A4 and MAGEA6). Integrative analysis with in vitro functional data and animal models provided supportive evidence for potential roles for these genetic aberrations in carcinogenesis. Pathway-based analysis of recurrently mutated genes recapitulated clustering in core signalling pathways in pancreatic ductal adenocarcinoma, and identified new mutated genes in each pathway. We also identified frequent and diverse somatic aberrations in genes described traditionally as embryonic regulators of axon guidance, particularly SLIT/ROBO signalling, which was also evident in murine Sleeping Beauty transposon-mediated somatic mutagenesis models of pancreatic cancer, providing further supportive evidence for the potential involvement of axon guidance genes in pancreatic carcinogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
196,21 € per year
only 3,85 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
In vivo CRISPR screens reveal SCAF1 and USP15 as drivers of pancreatic cancer
Article Open access 20 June 2024

The pancreatic cancer genome revisited
Article 04 June 2021
Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution
Article 13 January 2020
Accession codes
Data deposits
BAM files and associated metadata in XML format have been uploaded to the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega) under accession numbers GAS00001000154 and EGAS00001000343. Additional sequence data is located at dbGAP accession number phs000516.v1.p1.
Change history
References
- Jemal, A., Siegel, R., Xu, J. & Ward, E. Cancer statistics, 2010. CA Cancer J. Clin.60, 277–300 (2010) ArticleGoogle Scholar
- Butturini, G. et al. Influence of resection margins and treatment on survival in patients with pancreatic cancer: meta-analysis of randomized controlled trials. Arch. Surg.143, 75–83 (2008) ArticleGoogle Scholar
- Neoptolemos, J. P. et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. J. Am. Med. Assoc.304, 1073–1081 (2010) ArticleCASGoogle Scholar
- Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science321, 1801–1806 (2008) ArticleADSCASGoogle Scholar
- International Cancer Genome Consortium International network of cancer genome projects. Nature464, 993–998 (2010) ArticleADSGoogle Scholar
- Biankin, A. V. et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology137, 558–568 (2009) ArticleGoogle Scholar
- Chang, D. K. et al. Margin clearance and outcome in resected pancreatic cancer. J. Clin. Oncol.27, 2855–2862 (2009) ArticleGoogle Scholar
- Jamieson, N. B. et al. A prospective comparison of the prognostic value of tumor- and patient-related factors in patients undergoing potentially curative surgery for pancreatic ductal adenocarcinoma. Ann. Surg. Oncol.18, 2318–2328 (2011) ArticleGoogle Scholar
- Wang, L. et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res.22, 208–219 (2012) ArticleCASGoogle Scholar
- Song, S. et al. qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS ONE7, e45835 (2012) ArticleADSCASGoogle Scholar
- Samuel, N. & Hudson, T. J. The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma. Nature Rev. Gastroenterol. Hepatol.9, 77–87 (2012) ArticleCASGoogle Scholar
- Dees, N. D. et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Res.22, 1589–1598 (2012) ArticleCASGoogle Scholar
- Roberts, N. J. et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov.2, 41–46 (2011) ArticleGoogle Scholar
- Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol.12, R41 (2011) ArticleGoogle Scholar
- Sun, W. et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res.37, 5365–5377 (2009) ArticleCASGoogle Scholar
- Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature467, 1109–1113 (2010) ArticleADSCASGoogle Scholar
- Mann, K. M. et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA109, 5934–5941 (2012) ArticleADSCASGoogle Scholar
- Pérez-Mancera, P. A. et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature486, 266–270 (2012) ArticleGoogle Scholar
- Cheung, H. W. et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl Acad. Sci. USA108, 12372–12377 (2011) ArticleADSCASGoogle Scholar
- Mehlen, P., Delloye-Bourgeois, C. & Chedotal, A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nature Rev. Cancer11, 188–197 (2011) ArticleCASGoogle Scholar
- Sabatier, C. et al. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell117, 157–169 (2004) ArticleCASGoogle Scholar
- Trusolino, L. & Comoglio, P. M. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nature Rev. Cancer2, 289–300 (2002) ArticleCASGoogle Scholar
- Birnbaum, D. J. et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosom. Cancer50, 456–465 (2011) ArticleCASGoogle Scholar
- Bamford, S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer91, 355–358 (2004) ArticleCASGoogle Scholar
- Jones, S. et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mutat.33, 100–103 (2012) ArticleCASGoogle Scholar
- Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature469, 539–542 (2011) ArticleADSCASGoogle Scholar
- Comoglio, P. M. & Trusolino, L. Invasive growth: from development to metastasis. J. Clin. Invest.109, 857–862 (2002) ArticleCASGoogle Scholar
- Chédotal, A., Kerjan, G. & Moreau-Fauvarque, C. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ.12, 1044–1056 (2005) ArticleGoogle Scholar
- Ong, C. K. et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nature Genet.44, 690–693 (2012) ArticleCASGoogle Scholar
- Capparuccia, L. & Tamagnone, L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment–two sides of a coin. J. Cell Sci.122, 1723–1736 (2009) ArticleCASGoogle Scholar
- Müller, M. W. et al. Association of axon guidance factor semaphorin 3A with poor outcome in pancreatic cancer. Int. J. Cancer121, 2421–2433 (2007) ArticleGoogle Scholar
- Ellis, L. M. The role of neuropilins in cancer. Mol. Cancer Ther.5, 1099–1107 (2006) ArticleCASGoogle Scholar
- Kikuchi, K. et al. In vitro and in vivo characterization of a novel semaphorin 3A inhibitor, SM-216289 or xanthofulvin. J. Biol. Chem.278, 42985–42991 (2003) ArticleCASGoogle Scholar
- Cao, Y., DePinho, R. A., Ernst, M. & Vousden, K. Cancer research: past, present and future. Nature Rev. Cancer11, 749–754 (2011) ArticleCASGoogle Scholar
- Pajic, M., Scarlett, C. J., Chang, D. K., Sutherland, R. L. & Biankin, A. V. Preclinical strategies to define predictive biomarkers for therapeutically relevant cancer subtypes. Hum. Genet.130, 93–101 (2011) ArticleCASGoogle Scholar
- Biankin, A. V. & Hudson, T. J. Somatic variation and cancer: therapies lost in the mix. Hum. Genet.130, 79–91 (2011) ArticleGoogle Scholar
- Kris M. G., Meropol N. J., Winer E. P., eds. Accelerating Progress Against Cancer: ASCO’s Blueprint for Transforming Clinical and Translational Cancer Research (Am. Soc. Clin. Oncol., 2011)
- Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005) ArticleADSCASGoogle Scholar
Acknowledgements
This paper is dedicated to Robert L. Sutherland who died on 10 October 2012 of pancreatic cancer. We would like to thank C. Axford, D. Gwynne, M.-A. Brancato, S. Rowe, M. Thomas, S. Simpson and G. Hammond for central coordination of the Australian Pancreatic Cancer Genome Initiative, data management and quality control; M. Martyn-Smith, L. Braatvedt, H. Tang, V. Papangelis and M. Beilin for biospecimen acquisition; and W. Waterson, J. Shepperd, E. Campbell and E. Glasov for their efforts at the Queensland Centre for Medical Genomics. We also thank M. B. Hodgin, M. Debeljak and D. Trusty for technical assistance at Johns Hopkins University, and J. Lau, M. Karaus, K. Rabe, L. Zhang and T. Smyrk at the Mayo Clinic. We acknowledge the following funding support: National Health and Medical Research Council of Australia (NHMRC; 631701, 535903, 427601, 535914); Australian Government: Department of Innovation, Industry, Science, Research and Tertiary Education (DIISRTE); Australian Cancer Research Foundation (ACRF); Queensland Government (NIRAP); University of Queensland; Cancer Council NSW (SRP06-01; ICGC09-01; SRP11-01); Cancer Institute NSW (06/ECF/1-24, 09/CDF/2-40, 07/CDF/1-03, 10/CRF/1-01, 08/RSA/1-15, 07/CDF/1-28, 10/CDF/2-26,10/FRL/2-03, 06/RSA/1-05, 09/RIG/1-02, 10/TPG/1-04, 11/REG/1-10, 11/CDF/3-26); Garvan Institute of Medical Research; Avner Nahmani Pancreatic Cancer Research Foundation; R.T. Hall Trust; Petre Foundation; Jane Hemstritch in memory of Philip Hemstritch; Gastroenterological Society of Australia (GESA); American Association for Cancer Research (AACR) Landon Foundation – INNOVATOR Award; Royal Australasian College of Surgeons (RACS); Royal Australasian College of Physicians (RACP); Royal College of Pathologists of Australasia (RCPA); HGSC-BCM: NHGRI U54 HG003273; CPRIT grant RP101353-P7 (Tumor Banking for Genomic Research and Clinical Translation Site 1); The Ontario Institute for Cancer Research; The Ontario Ministry of Economic Development and Innovation; Canada Foundation for Innovation; Pancreatic Cancer Genetic Epidemiology Consortium, NIH grant R01 CA97075; The Agency for Science, Technology, and Research (Singapore); University of Verona and Italian Ministry of University (FIRB RBAP10AHJB), Rome, Italy; Cancer Research UK; Wellcome Trust; CPRIT (Cancer Prevention Research Institute of Texas); NIH P50CA062924 (SPORE) and P01CA134292 (PPG); The Sol Goldman Pancreatic Cancer Research Center; NCI grant P50 CA102701 (Mayo Clinic SPORE in Pancreatic Cancer) and NCI grant R01 CA97075 (Pancreatic Cancer Genetic Epidemiology Consortium); NIH SPORE grant 2P50CA101955 (UMN/UAB), and AIRC (Associazione Italiana Ricerca sul Cancro) 5xmille grant 12182, Italy.
Author information
- Robert L. Sutherland: Deceased.
Authors and Affiliations
- The Kinghorn Cancer Centre, 370 Victoria Street, Darlinghurst, and the Cancer Research Program, Garvan Institute of Medical Research, 384 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia., Andrew V. Biankin, Amber L. Johns, Jianmin Wu, David K. Chang, Mark J. Cowley, Marina Pajic, Christopher J. Scarlett, Anthony J. Gill, Andreia V. Pinho, Ilse Rooman, Mark Pinese, R. Scott Mead, Jeremy L. Humphris, Warren Kaplan, Marc D. Jones, Emily K. Colvin, Adnan M. Nagrial, Emily S. Humphrey, Angela Chou, Venessa T. Chin, Lorraine A. Chantrill, Amanda Mawson, James G. Kench, Jessica A. Lovell, Roger J. Daly, Christopher Toon, Elizabeth A. Musgrove & Robert L. Sutherland
- Department of Surgery, Bankstown Hospital, Eldridge Road, Bankstown, Sydney, New South Wales 2200, Australia., Andrew V. Biankin, David K. Chang & Neil D. Merrett
- South Western Sydney Clinical School, Faculty of Medicine, University of NewSouth Wales, Liverpool, 2170, NewSouth Wales, Australia Andrew V. Biankin & David K. Chang
- Queensland Centre for Medical Genomics, Institute for Molecular Bioscience, The University of Queensland, St Lucia, Brisbane, 4072, Queensland, Australia Nicola Waddell, Karin S. Kassahn, David K. Miller, Peter J. Wilson, Ann-Marie Patch, Brooke B. Gardiner, Sarah Song, Ivon Harliwong, Senel Idrisoglu, Craig Nourse, Ehsan Nourbakhsh, Suzanne Manning, Shivangi Wani, Milena Gongora, Matthew Anderson, Oliver Holmes, Conrad Leonard, Darrin Taylor, Scott Wood, Qinying Xu, Katia Nones, J. Lynn Fink, Angelika Christ, Tim Bruxner, Nicole Cloonan, Felicity Newell, John V. Pearson & Sean M. Grimmond
- Department of Molecular and Human Genetics, Human Genome Sequencing Center, Baylor College of Medicine, One Baylor Plaza, MS226, Houston, Texas 77030-3411, USA., Marie-Claude Gingras, Nipun Kakkar, Fengmei Zhao, Yuan Qing Wu, Min Wang, Donna M. Muzny, Jeffrey G. Reid, Jennifer Drummond, Kyle Chang, Yi Han, Lora R. Lewis, Huyen Dinh, Christian J. Buhay, David A. Wheeler & Richard A. Gibbs
- Michael E. DeBakey Department of Surgery, Baylor College of Medicine, One Baylor Plaza, Houston, Texas 77030-3411, USA., Marie-Claude Gingras, William E. Fisher & Sally E. Hodges
- Ontario Institute for Cancer Research, Toronto, M5G 0A3, Canada Lakshmi B. Muthuswamy, Timothy Beck, Lee Timms, Michelle Sam, Kimberly Begley, Andrew Brown, Deepa Pai, Ami Panchal, Nicholas Buchner, Richard De Borja, Robert E. Denroche, Christina K. Yung, Nicole Onetto, Lincoln D. Stein, Thomas J. Hudson & John D. McPherson
- School of Environmental & Life Sciences, University of Newcastle, Ourimbah, 2258, New South Wales, Australia Christopher J. Scarlett
- Department of Anatomical Pathology, Royal North Shore Hospital, St Leonards, Sydney, 2065, New South Wales, Australia Anthony J. Gill & Jaswinder S. Samra
- University of Sydney, Sydney, 2006, New South Wales, Australia Anthony J. Gill & James G. Kench
- Life Technologies, Brisbane, 4000, Queensland, Australia Gabriel Kolle
- Department of Anatomical Pathology, Sydpath, St Vincent’s Hospital, Sydney, 2010, New South Wales, Australia R. Scott Mead & Angela Chou
- Department of Surgery, Royal North Shore Hospital, St Leonards, Sydney, 2065, New South Wales, Australia Jaswinder S. Samra
- Tissue Pathology and Diagnostic Oncology, Royal Prince Alfred Hospital, Camperdown, 2050, New South Wales, Australia James G. Kench
- School of Medicine, University of Western Sydney, Penrith, 2175, New South Wales, Australia Neil D. Merrett
- Department of Surgery, Fremantle Hospital, Alma Street, Fremantle, Western Australia 6160, Australia., Krishna Epari
- Department of Gastroenterology, Royal Adelaide Hospital, North Terrace, Adelaide, South Australia 5000, Australia., Nam Q. Nguyen
- Department of Surgery, The University of Queensland, Princess Alexandra Hospital, Ipswich Road, Woollongabba, Queensland 4102, Australia., Andrew Barbour
- School of Surgery M507, University of Western Australia, 35 Stirling Hwy, Nedlands 6009, Australia and St John of God Pathology, 12 Salvado Road, Subiaco, Western Australia 6008, USA., Nikolajs Zeps
- The Elkins Pancreas Center atBaylor College ofMedicine, 6620 Main Street, Suite 1450, Houston, Texas 77030, USA., William E. Fisher & Sally E. Hodges
- David Geffen School of Medicine at UCLA, Los Angeles, 90024, Callfornia, USA F. Charles Brunicardi
- University Health Network, Toronto, M5G2C4, Canada Stefano Serra, Ming-Sound Tsao, Patricia A. Shaw & Steven Gallinger
- Mayo Clinic, Rochester, 55905, Minnesota, USA Debabrata Mukhopadhyay & Gloria M. Petersen
- Mount Sinai Hospital, Toronto, M5G 1X5, Canada Steven Gallinger
- Department of Pathology, The Sol Goldman Pancreatic Cancer Research Center, the Johns Hopkins University School of Medicine, Baltimore, 21231, Maryland, USA Ralph H. Hruban, Anirban Maitra, Christine A. Iacobuzio-Donahue, Richard A. Morgan & James R. Eshleman
- Department of Surgery, The Sol Goldman Pancreatic Cancer Research Center, the Johns Hopkins University School of Medicine, Baltimore, 21231, Maryland, USA Richard D. Schulick & Christopher L. Wolfgang
- ARC-NET center for applied research on cancer, University and Hospital Trust of Verona, Verona, 37134, Italy Rita T. Lawlor, Vincenzo Corbo & Aldo Scarpa
- Department of Pathology and Diagnostics, University of Veron, Verona, 37134, Italy Paola Capelli, Maria Scardoni & Aldo Scarpa
- Department of Surgery and Oncology, University and Hospital Trust of Verona, Verona, 37134, Italy Giampaolo Tortora
- Division of Hematology and Oncology, University of California, San Francisco, 94115, USA Margaret A. Tempero
- Cancer Research Program, The Methodist Hospital Research Institute, 6670 Bertner Avenue, Houston, Texas 77030, USA., Karen M. Mann, Nancy A. Jenkins & Neal G. Copeland
- and Department of Oncology, Li Ka Shing Centre, Cambridge Research Institute, Cancer Research UK, Robinson Way, CB2 0RE, Cambridge, UK Pedro A. Perez-Mancera & David A. Tuveson
- Experimental Cancer Genetics, Wellcome Trust Sanger Institute, Hinxton, CB10 1HH, UK David J. Adams & Alistair G. Rust
- Masonic Cancer Center, University of Minnesota, Minneapolis, 55455, Minnesota, USA David A. Largaespada
- Bioinformatics and Statistics, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands., Lodewyk F. A. Wessels
- St Vincent’s Clinical School, Faculty of Medicine, University of New South Wales, New South Wales, 2010, Australia Elizabeth A. Musgrove & Robert L. Sutherland
- The Kinghorn Cancer Centre, Cancer Research Program, Garvan Institute of Medical Research, 370 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia., Andrew V. Biankin, Amber L. Johns, Amanda Mawson, David K. Chang, Christopher J. Scarlett, Mary-Anne L. Brancato, Sarah J. Rowe, Skye L. Simpson, Mona Martyn-Smith, Michelle T. Thomas, Lorraine A. Chantrill, Venessa T. Chin, Angela Chou, Mark J. Cowley, Jeremy L. Humphris, Marc D. Jones, R. Scott Mead, Adnan M. Nagrial, Marina Pajic, Jessica Pettit, Mark Pinese, Ilse Rooman, Jianmin Wu, Jiang Tao, Renee DiPietro, Clare Watson, Rachel Wong, Andreia V. Pinho, Marc Giry-Laterriere, Roger J. Daly, Elizabeth A. Musgrove & Robert L. Sutherland
- Queensland Center for Medical Genomics, Institute for Molecular Bioscience, University of Queensland, St Lucia, 4072, Queensland, Australia Sean M. Grimmond, Nicola Waddell, Karin S. Kassahn, David K. Miller, Peter J. Wilson, Ann-Marie Patch, Sarah Song, Ivon Harliwong, Senel Idrisoglu, Craig Nourse, Ehsan Nourbakhsh, Suzanne Manning, Shivangi Wani, Milena Gongora, Matthew Anderson, Oliver Holmes, Conrad Leonard, Darrin Taylor, Scott Wood, Qinying Xu, Katia Nones, J. Lynn Fink, Angelika Christ, Tim Bruxner, Nicole Cloonan, Felicity Newell & John V. Pearson
- Royal North Shore Hospital, Westbourne Street,, St Leonards, 2065, New South Wales, Australia Jaswinder S. Samra, Anthony J. Gill, Nick Pavlakis, Alex Guminski & Christopher Toon
- Bankstown Hospital, Eldridge Road,, Bankstown, 2200, New South Wales, Australia Andrew V. Biankin, Ray Asghari, Neil D. Merrett, David K. Chang, Darren A. Pavey & Amitabha Das
- Liverpool Hospital, Elizabeth Street, Liverpool, 2170, New South Wales, Australia Peter H. Cosman, Kasim Ismail & Chelsie O’Connor
- Westmead Hospital, Hawkesbury and Darcy Roads, Westmead, New South Wales 2145, Australia., Vincent W. Lam, Duncan McLeod, Henry C. Pleass, Arthur Richardson & Virginia James
- Royal Prince Alfred Hospital, Missenden Road, Camperdown, New South Wales 2050, Australia., James G. Kench, Caroline L. Cooper, David Joseph, Charbel Sandroussi, Michael Crawford & James Gallagher
- Fremantle Hospital, Alma Street, Fremantle, Western Australia 6959, Australia., Michael Texler, Cindy Forrest, Andrew Laycock, Krishna P. Epari, Mo Ballal, David R. Fletcher & Sanjay Mukhedkar
- Sir Charles Gairdner Hospital, Hospital Avenue, Nedlands, 6009, Western Australia, Australia Nigel A. Spry, Bastiaan DeBoer & Ming Chai
- St John of God Healthcare, 12 Salvado Road, Subiaco, Western Australia 6008, Australia., Nikolajs Zeps, Maria Beilin & Kynan Feeney
- Royal Adelaide Hospital, North Terrace, Adelaide, 5000, South Australia, Australia Nam Q. Nguyen, Andrew R. Ruszkiewicz, Chris Worthley, Chuan P. Tan & Tamara Debrencini
- Flinders Medical Centre, Flinders Drive, Bedford Park, 5042, South Australia, Australia John Chen, Mark E. Brooke-Smith & Virginia Papangelis
- Greenslopes Private Hospital, Newdegate Street, Greenslopes, Queensland 4120, Australia., Henry Tang & Andrew P. Barbour
- Envoi Pathology, 1/49 Butterfield Street, Herston, Queensland 4006, Australia., Andrew D. Clouston & Patrick Martin
- Princess Alexandria Hospital, Cornwall Street & Ipswich Road, Woolloongabba, Queensland 4102, Australia., Thomas J. O’Rourke, Amy Chiang, Jonathan W. Fawcett, Kellee Slater, Shinn Yeung, Michael Hatzifotis & Peter Hodgkinson
- Austin Hospital, 145 Studley Road, Heidelberg, Victoria 3084, Australia., Christopher Christophi, Mehrdad Nikfarjam & Angela Mountain Victorian Cancer Biobank
- Victorian Cancer Biobank, 1 Rathdowne Street, Carlton, Victoria 3053, Australia., Angela Mountain Victorian Cancer Biobank
- Johns Hopkins Medical Institutes, 600 North Wolfe Street, Baltimore, Maryland 21287, USA., James R. Eshleman, Ralph H. Hruban, Anirban Maitra, Christine A. Iacobuzio-Donahue, Richard D. Schulick, Christopher L. Wolfgang, Richard A. Morgan & Mary B. Hodgin
- ARC-NET Center for Applied Research on Cancer, University of Verona, Via dell’Artigliere, 19 37129 Verona 37134, Italy., Aldo Scarpa, Rita T. Lawlor, Paola Capelli, Stefania Beghelli, Vincenzo Corbo, Maria Scardoni, Paolo Pederzoli, Giampaolo Tortora & Claudio Bassi
- University of California, San Francisco, 1600 Divisadero Street, San Francisco, California 94115, USA., Margaret A. Tempero
- Andrew V. Biankin







